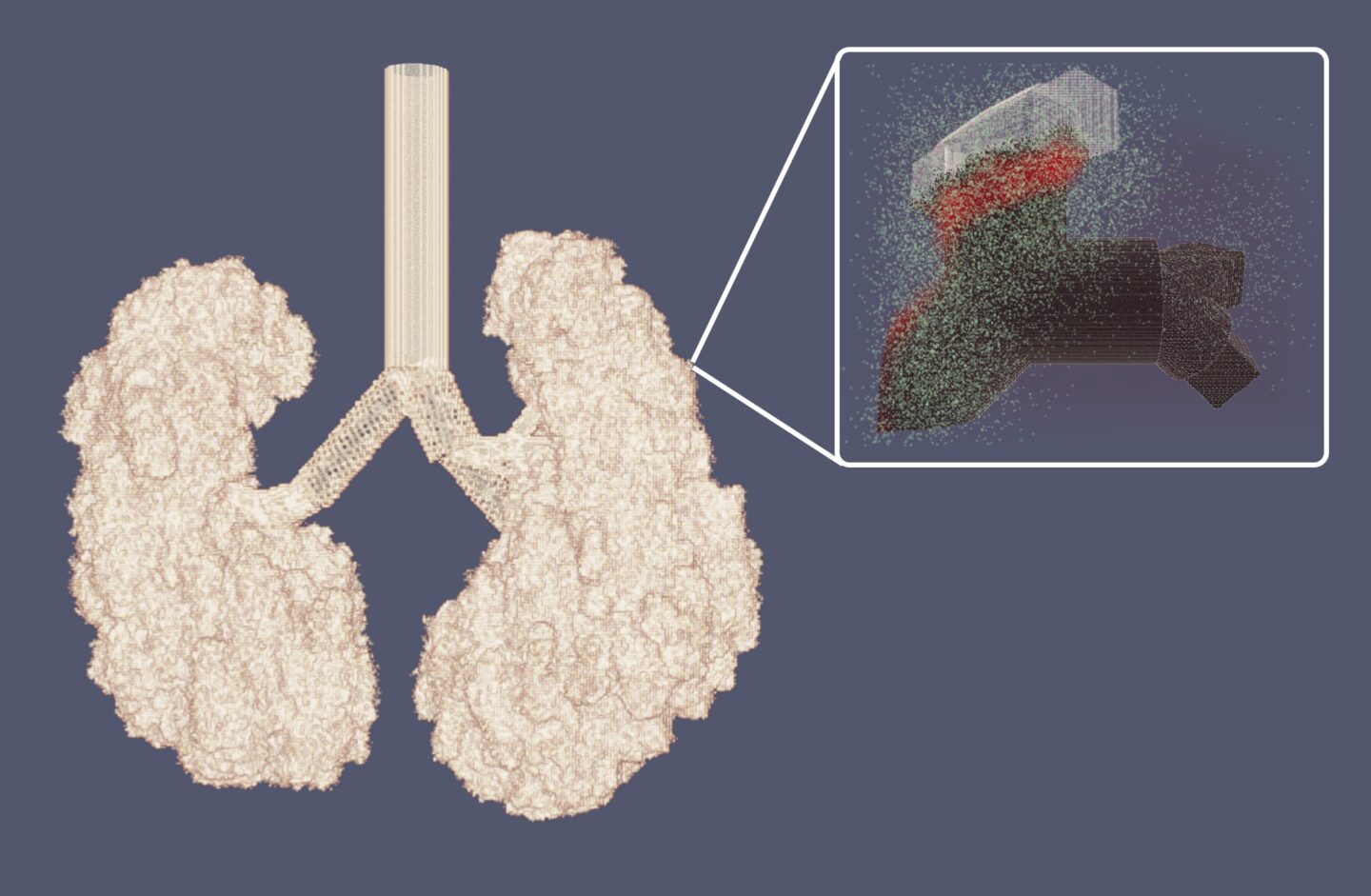
Lawrence Berkeley National Laboratory (Berkeley Lab) computational research scientist Steve Hofmeyr and a team at the University of New Mexico and Arizona State University have succeeded in modeling how COVID-19 and other viral infections spread in the lungs, capturing a cell-by-cell process as it’s never been captured before.
Their new computational model, Spatial Immune Model of Coronavirus (SIMCoV), was developed using the Cori supercomputer at the National Energy Research Scientific Computing Center (NERSC), a U.S. Department of Energy user facility located at Berkeley Lab. SIMCoV offers a 3D simulation of a portion of the lung and shows how viral infections can spread at the cellular level. Specifically, it shows how T-cells – which fight viruses inside our cells at the same time that B-cells, or antibodies, fight viruses between the cells – interact with viral load and other variables to influence the progression of an infection. The model treats each of the billions of epithelial cells lining the lungs and the hundreds of millions of T-cells defending them as an individual agent, a level of detail made possible by using significant computing resources.
“We feel this model has a lot of potential to explain various aspects of [COVID-19], and computational modeling in this case is very powerful because it’s extremely difficult to image and understand what’s going on at this level of the immune system response,” said Hofmeyr, a co-author on a paper outlining this work published in PLOS Computational Biology in December. “Even measuring levels of T-cells is not easy; we can measure levels of T-cells in the blood, but that doesn’t necessarily indicate how many T-cells have gone into the lungs themselves to fight infections. This model includes a lot of scope for understanding that.”
Previous models based on differential equations portray the lungs as uniform and assume that the growth of viral infections is also distributed evenly within the lungs. This new cell-by-cell model offers much more spatial detail, showing that infections may be active in some areas and dormant in others, as well as how different areas might flare up and then recede over time as the virus and the immune system interact.
According to Hofmeyr, this model also takes into account the spatial distribution of the virus. “We’ve done experiments that show that, for example, you have a certain number of virions, or encapsulated viruses, in the initial infection,” he said. “If you take a fixed number of them and concentrate them in one location compared with taking that same number and distributing them through the lungs, the second case is much, much worse. And the reason it’s much, much worse is that the immune system has to find the different infections, and the T-cells have to be distributed to find the infection in the different locations, which is a much more challenging problem.”
UPC++ Library Lends Key Support
Hofmeyr and his colleagues developed the model using UPC++, a distributed memory library for C++ developed by the Pagoda Project at Berkeley Lab, an Exascale Computing Project-funded endeavor that researches and develops cutting-edge software for implementing high-performance applications and software infrastructure using the Partitioned Global Address model. UPC++ supports physically distributed global memory, allowing it to scale effectively to large numbers of processors, including a possible future SIMCoV model of the complete lung.
“I’ve been using UPC++ a lot lately and finding that it’s an awesome programming model. It’s very nicely done and very simple to parallelize things,” Hofmeyr said. “It has some really powerful features.”
One possible extension of SIMCoV is as a method of modeling the mechanism by which the virus actually spreads deep within the lungs, whether it’s spread from one part of the lobe to another through air circulation with each inhalation and exhalation, or spreads repeatedly to the lower lung from an infection in the upper respiratory tract. The same model may also be of use in understanding the mechanism behind “long COVID,” in which those infected with COVID continue to experience symptoms for a longer-than-normal period of time.
The SIMCoV model was developed in the context of COVID-19, but its findings can be applied to other viral lung infections as well. Hofmeyr hopes that, in the future, the model might be applied to influenza, which kills hundreds of thousands of people around the world each year. The model may also be of use in steering research on interventions for viral lung infections; by experimenting with different interventions and their effects at different times in a disease’s progression, scientists may be able to identify courses of treatment that are likely to succeed.
Hofmeyr compares the size and power of the SIMCoV model in its current form to those typically applied to less human-scaled situations: “These high-powered models are similar to the kind of modeling you would do for physical systems or chemical systems,” he said, “where you can’t really do the experiments you really need to do because they’re just too massive, and you don’t know how to answer all the questions. You can’t measure everything; maybe you’re doing experiments in fusion science or something. This kind of model provides a similar sort of assistance.”
About Computing Sciences at Berkeley Lab
High performance computing plays a critical role in scientific discovery. Researchers increasingly rely on advances in computer science, mathematics, computational science, data science, and large-scale computing and networking to increase our understanding of ourselves, our planet, and our universe. Berkeley Lab's Computing Sciences Area researches, develops, and deploys new foundations, tools, and technologies to meet these needs and to advance research across a broad range of scientific disciplines.