Building on Berkeley Lab’s rich tradition of team science, we combine deep expertise in mathematics, statistics, computing, and data sciences with a wide range of scientific disciplines to drive AI innovation. We collaborate with researchers across domains to generate, process, and curate vast datasets, which serve as critical resources for scientific discovery.
In parallel, we develop and deploy cutting-edge AI models and tools. By adapting existing machine learning methods and creating new ones, we address the diverse, evolving needs of scientific research, answering fundamental questions and enabling breakthroughs across various fields. These AI models are applied to solve complex scientific problems and accelerate discovery in key research areas.
Our efforts are supported by robust high-performance computing and networking infrastructure. We operate the Department of Energy’s National Energy Research Scientific Computing Center (NERSC) and the Energy Sciences Network (ESnet), which provide the computational power, data storage, and remote access necessary for AI training, large-scale simulations, and collaborative research at distant experimental facilities.
Our Research Pillars:
- New Models and Methods
- Learning for Scientific Discovery
- Supercomputing-Scale AI
- Secure Machine Learning & ML for Security
- Statistics and Applied Mathematics
- Data and Science Informed Learning

gpCAM is an open-source software tool that uses artificial intelligence to help scientists automatically plan, collect, and analyze data from experiments and simulations, making research faster and more efficient.

A deep learning system that automates the analysis of plant root images, providing researchers with fast, precise insights to advance agricultural and environmental science.
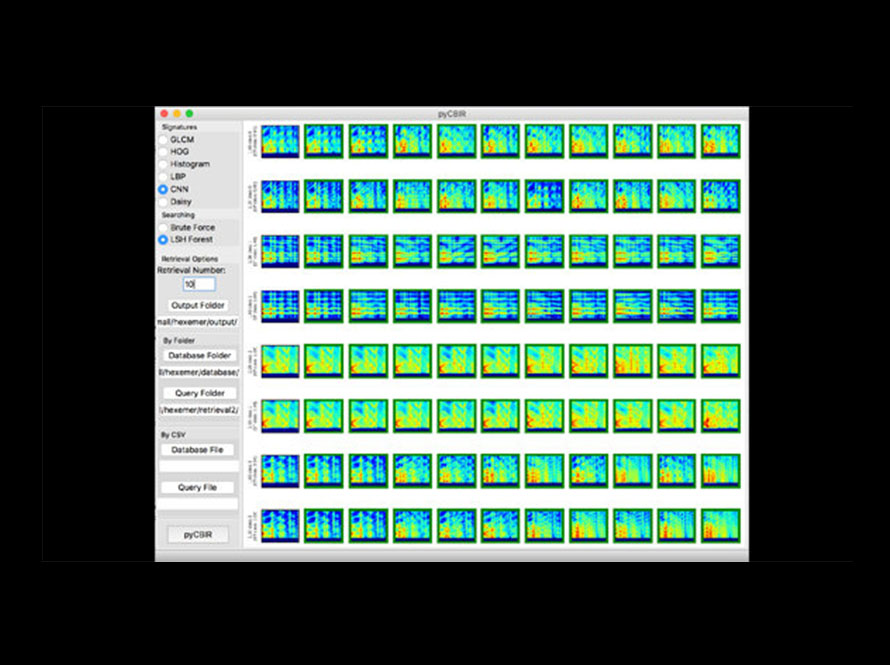
A deep learning-based image search tool that allows scientists to find and compare scientific images across large datasets, enhancing data exploration and discovery.

CAMERA develops and maintains a suite of open-source AI and machine learning software—including pyMSDtorch, MSDNet, and other advanced tools—that enable autonomous experimentation, advanced data analysis, and image reconstruction across scientific domains.

The ENDURABLE project develops advanced tools for robust data aggregation and deep learning model training, making it easier for scientists to build complex machine learning datasets and harness the full potential of AI for scientific discovery.
FunFact is a Python package that automates matrix and tensor factorization algorithms, supporting applications such as neural network compression and quantum circuit synthesis, and is built on modern machine learning frameworks for scalable, flexible scientific computing.
GPTune is an AI-driven autotuning tool that leverages Bayesian optimization and transfer learning to efficiently solve complex black-box optimization problems in scientific computing.
GraphDot is a GPU-accelerated library for machine learning on graphs, enabling fast and customizable graph similarity computations using advanced kernel methods and graph convolution techniques for scientific data analysis.

Scalable Graph Learning for Scientific Discovery develops efficient, distributed-memory algorithms and methods to enable large-scale, memory-efficient graph representation learning (GRL) and hypergraph learning for advancing research in fields such as structural biology, computational chemistry, and particle physics.
This project investigates the unique performance characteristics of AI training and inference compared to traditional HPC applications, focusing on how computational methods, precision, and specialized hardware architectures impact efficiency and scalability. By analyzing the interactions between scientific workloads, AI frameworks, and hardware, the project aims to identify bottlenecks and guide the optimization of current and future AI systems.

pyAMReX provides GPU-enabled Python bindings for the AMReX mesh-refinement framework, allowing seamless integration of AI and machine learning models with large-scale scientific simulations for in situ analysis and rapid prototyping.

AI-Accelerated Astrophysical Reaction Networks use machine learning to replace computationally intensive reaction network solvers in astrophysical simulations, dramatically speeding up calculations while maintaining scientific accuracy.

Data-Driven Modeling of Complex Fluids combines machine learning with multiscale modeling frameworks to predict the behavior of complex, microstructural fluids, enhancing the accuracy and efficiency of simulations.
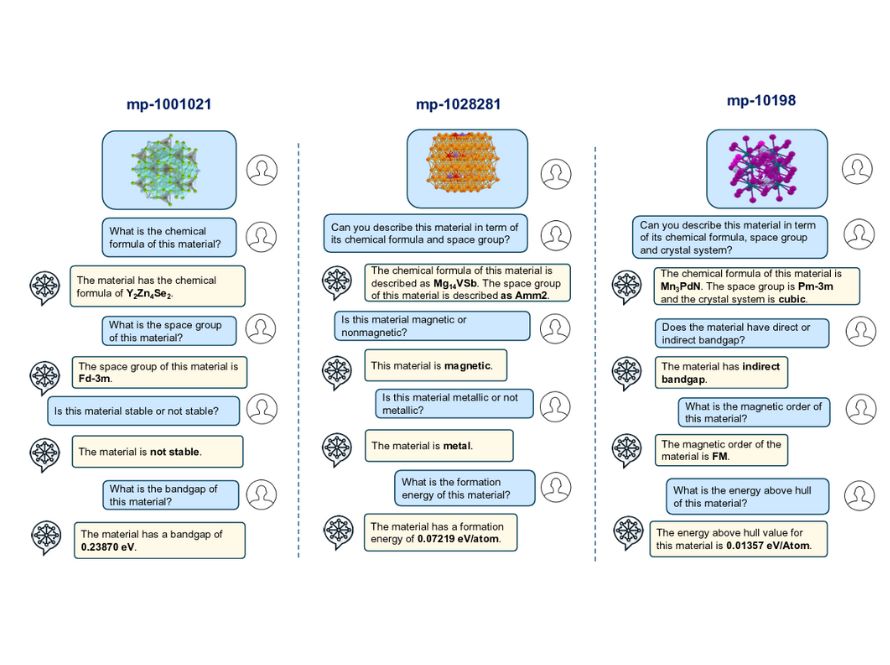
The ManyChat multimodal large language model for materials discovery develops advanced language models tailored for materials science, integrating diverse data types to accelerate materials analysis, prediction, and design.

INDIE (Intelligent Distribution for Advanced Wireless Networks with Scientific Data Microservices) is a software platform that provides intelligent data management, computational task composition, and workflow coordination for scientific applications across advanced wireless networks like 5G and 6G.

The Vertically Integrated Artificial Intelligence for Sensing and High-Performance Computing (VIAS) project develops next-generation detectors that use vertically integrated, chiplet-based circuits and advanced AI hardware to enable fast, efficient data analysis in challenging environments.

Berkeley Lab collaborates with Meta to co-lead the development of Open Molecules 2025, an unprecedented dataset of over 100 million 3D molecular simulations that aims to revolutionize machine learning tools for accurately modeling complex chemical reactions, thereby transforming research in materials science, biology, and energy technologies.

AMCR’s Silvia Crevelli serves as the principal investigator of a project that develops scalable AI methods in collaboration with the VA, leveraging billions of clinical records to create advanced clinical risk models that improve precision medicine and public health outcomes, including suicide risk prediction and treatment efficacy in lung cancer, by integrating structured data from Electronic Health Records and utilizing large language models for comprehensive analysis.