Biological
Biological systems are dynamical systems that operate over huge ranges of time and length scales. Understanding these systems is often greatly aided by the use of simulations, powered by high performance computers (HPC) to span these spatio-temporal scales in ways that can not be done experimentally. Furthermore, formulating precise mathematical models underlying biological phenomena often brings insight in and of itself.
Burgeoning research at the intersection of applied math and biology at Berkeley Lab uses HPC systems to simulate the biophysics of electrical signal propagation in the brain, leverages parallel codes to understand the spread of SARS-CoV-2 in the lungs, and is exploring new mathematical challenges at the biophysical interface of solution composition and membranes, fluid dynamics in bioreactors.
Projects
ExaEpi
Researchers are working on two projects funded by the DOE Bio-preparedness Research Virtual Environment (BRaVE) Initiative on next-generation epidemiological modeling. At the heart of both efforts is ExaEpi, the new agent-based epidemiological modeling code based on AMReX. ExaEpi allows us to run large ensembles very quickly at scale using DOE's HPC resources. This will allow us to not only calibrate the models for a disease like Covid-19 using data on wastewater, hospitalizations, deaths, etc, but we can also use it to understand both the degeneracies and importance of the underlying parameters in agent-based models. Contact: Peter Nugent
SIMCoV
Spatial Immune Model of Coronavirus (SIMCoV) is a scalable computational model written in UPC++ that simulates hundreds of millions of lung cells, including respiratory epithelial cells and T cells. SIMCoV replicates viral growth dynamics observed in patients and shows that spatially dispersed infections lead to increased viral loads. The model shows how the timing and strength of the T-cell response can affect viral persistence, oscillations, and control. By incorporating spatial interactions, SIMCoV provides a parsimonious explanation for the dramatically different viral load trajectories among patients by varying only the number of initial sites of infection and the magnitude and timing of the T-cell immune response. Contact: Steven Hofmeyr (Hofmeyr on the Web)
Brain Biophysics: Bridging spatiotemporal scales in the brain with multi-modal data and biophysical simulations
Biological systems are organized and function across a large range of spatial and temporal scales. One of the most complicated, multi-scale biological systems is the brain, in which myriad neurons are organized into microcircuits (or “columns”) that perform specific computations but are simultaneously integrated into larger networks. How local neural processing is organized and coordinated in the context of broader neural networks – and how this is reflected in spiking activity and different frequency bands – is poorly understood. We use large-scale biophysically detailed simulations to bridge spatiotemporal scales in the nervous system. Contact: Kristofer Bouchard
News
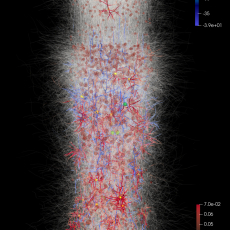
Neuroscience Simulations at NERSC Shed Light on Origins of Human Brain Recordings
Using simulations run at NERSC, a team of researchers at Berkeley Lab has found the origin of cortical surface electrical signals in the brain and discovered why the signals originate where they do. Read More »
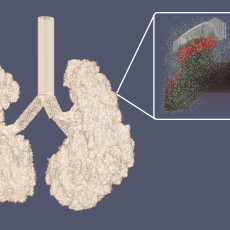
SIMCoV Models Cell-by-Cell Spread of Respiratory Infections
Berkeley Lab computational research scientist Steve Hofmeyr is part of a team that developed SimCoV, which can provide a 3D simulation of a portion of the lung and model the ways in which COVID-19 and other viral infections spread in the lungs. Read More »

Summer Students Use AMReX to Study Cancer Cell Migration
As part of Berkeley Lab's 2021 Computing Sciences Summer Program, a team of undergrads led by senior scientist Ann Almgren got a little closer to understanding how groups of cancer cells may respond to different kinds of forces. Read More »







 Instagram
Instagram YouTube
YouTube